Services/Equipment
Illumina sequencing platforms
HiSeq 1000

NextSeq 500

Our facility offers a full range of services for library construction and sequencing with the Illumina HiSeq 1000 and NextSeq 500 sequencers.
We have extensive experience in transcriptome sequencing (for expression profiling and transcritome assembly), genome requencing and targeted resequencing and other applications using RNAseq and DNAseq.
Sequencing options
| Single-read or Paired-end | Fragments can be sequenced from one end (single reads) or from both ends (paired-end reads). The former option is used for gene expression profiling, the latter for a more in depth characterization of transcripts isoforms, novel genes etc... |
| Read length | Recommended length for DNA-Seq, RNA-Seq sequencing is 75nt-150nt. |
| Indexing | All libraries, except from mate-pair libraries, are individually barcoded in order to allow the multiplexing of multiple libraries in a single lane. |
| Yields | HiSeq 1000: Typical yields are 140-200 millions of fragments sequenced per lane. This means that from 14Gb to 20Gb of data are generated for single reads (100nt) and from 28 to 40 Gb of data are generated for paired-end reads (100nt X 2). NextSeq 500: Typical yields are 400 millions of fragments sequenced per flowcell (High Output). This means that about 30Gb of data are generated for single reads (75nt) and 120Gb of data are generated for paired-end reads (150nt X 2). |
For prices of our sequencing and library production services see our pricing page. For informations on sample requirements see our protocols page.
MiSeq

For smaller projects and/or longer sequences we have access to an Illumina MiSeq Personal Sequencer. MiSeq sequencer produces up to 8.5 Gb of data from 2 x 250 paired-end reads in a single run.
Oxford Nanopore MinION

In 2014 Functional Genomics Lab has been selected from Oxford Nanopore to develop and test innovative applications using the third generation sequencing platform MinION in the frame of the MinION Access Program.
Pacific Biosciences

For de novo genome assembly applications we have access to Pacific Biosciences single molecule real time sequencing technologies which allows to reach an average read length of 10,000 - 15,000 bases, with the longest reads exceeding 40,000 bases.
Other instrumentation
Illumina BeadXpress
The BeadXpress system from Illumina offers a cost effective platform for assaying 48 to 384 SNPs in essentially any number of individuals. The BeadXpress platform primarily uses the Golden Gate SNP genotyping assay.
Covaris S220

Uses Covaris AFA technology to shear DNA. Double stranded DNA (unlike RNA) will shear when exposed to the energy of AFA. This shearing is site independent and is a perfect substrate for the production of DNA libraries. The system is computer controlled and once the conditions have been optimised for the size range you require it is highly repeatable. In addition to this AFA allows for a large number of samples to be processed in a short period of time with no washing between samples.
- Allows highly reproducible results
- Temperature controlled process
- Generate tight frament distributions
Stratagene MX3000P Real Time PCR

The Mx3000P RT-PCR system has a scanning PMT (350-700 nm) and a thermal cycler with a temperature range of 25-95°C at ± 0.25°C variation in sample temperature across a 96-well plate. The Mx3000P is specifically designed to ensure optimal results with multiplex assays using up to four targets in the same sample. In addition, the Mx3000P achieves linearity of amplification over seven orders of magnitude and possess the ability to detect two-fold differences in initial template quantity with 99.7% confidence.
Real Time PCR is used to measure the concentration of libraries against a standard curve before sequencing. Used also to validate relative expression changes detected by RNA-Seq analysis.
Nanodrop 1000

Allows to perform spectrophotometric measures of 1 ul samples in the full spectrum (220nm-750nm). It's used to check sample concentration and purity. The following parameters are determined:
- A260/A280 ratio (above 1.8)
- A260/A230 ratio (between 1.8-2.1)
Nanodrop 3300

Nanodrop 3300 is a spectrofluorimeter which allows to determine the concentration of 1 ul RNA or DNA samples.
Agilent 2100 Bioanalyzer

The Agilent Bioanalyzer RNA assay leverages microfluidics technology, enabling the quality analysis of RNA using only 1ul of sample. The assay is run on the Agilent 2100 Bioanalyzer instrument and utilises the Agilent 2100 Expert Software to analyze and display results. An RNA Integrity Number (RIN score) is generated for each sample on a scale of 1-10 (1=lowest; 10=highest) as an indication of RNA quality. The 18s/28s ratio and an estimation of concentration is also produced.
Sage Science Pippin Prep
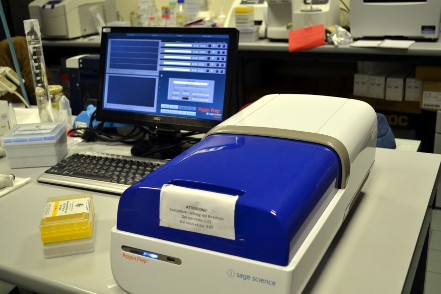
The Pippin Prep is a preparative electrophoresis platform that separates and extracts DNA fragments. Using pre-cast and disposable gel cassettes, DNA is automatically collected in buffer according to software-input size ranges. DNA fractions (typically adapter-ligate NGS libraries) are removed using standard pipettes. Workflow requires about 1-2 minutes of hands on time per sample, and cassettes are available for extractions between 50 bp and 8kb. Typical run times (four samples per run) are between 50 and 100 minutes.
Functional Genomics Lab
Department of Biotechnologies, University of Verona
Strada le Grazie, 15 - Cà Vignal, 1 - 37134 (VR) - Italy
Phone: +39 045 802 7058
Fax: +39 045 802 7929